Research outcomes of the Program for the Advancement of Next Generation Research Projects
Faculty of Pharmacy
Fumihito Ueda
DNA methylation is a typical epigenetic modification of DNA that negatively regulates gene expression. It is responsible for the mode of gene expression during development and differentiation, and is also associated with aging and diseases such as cancer. While DNA methylation is globally repressed in cancer cells, it is enhanced in certain regions, such as CpG island in promoters and enhancers of genes related to DNA damage repair or tumor suppression, and thought to promote biological alterations on a cellular level. However, the underlying molecular basis of how cancer cells induce low DNA methylation and aberrant DNA methylation in specific regions is largely unknown.
It has been known that aberrant DNA methylation is induced in normal tissues in the very early stages of cancer development, and the degree of methylation accumulation in normal tissues is correlated with cancer risk. Recently, it was demonstrated that chronic inflammation induced the suppression of erasers (TET methylcytosine dioxygenases [TETs]) and the activation of writers (DNA methyltransferases [DNMTs]) of DNA methylation, causing synergistically induced aberrant DNA methylation. However, the mechanisms by which methylation is selectively induced in specific DNA regions remain enigmatic.
Anaplastic large cell lymphoma (ALCL) is a non-Hodgkin's lymphoma of T cell origin in which NPM-ALK fusion protein is detected in more than 30% of patients as a result of chromosomal translocations. It has been shown that NPM-ALK promotes DNA methylation in the promoter regions of various genes and silences them. However, analysis of these gene expressions in multiple cell lines derived from patients with ALCL revealed that there are regions with different methylation states and that they are not necessarily silenced in the same way. Our team thoroughly investigated the differences between the cell lines and analyzed the DNA methylation status in detail to elucidate the factors that determine DNA methylation patterns in ALCL distinct from NPM-ALK.
Comprehensive analysis of gene expression induced by treatment of DNA demethylating agent 5-Aza by RNA-Seq analysis and database analysis of gene expression or methylation data in various cell lines derived from patients with ALCL revealed some candidate genes that may influence DNA methylation pattern in ALCL cells. Among them, we found that STAT1 expression at the protein level or STAT1 activation level was inversely-correlated with DNA methylation levels in the promoter of specific genes in ALCL cells. Furthermore, we clarified that STAT1 activation by IFN-γ stimulation triggered interaction of STAT1 with TET2, causing DNA demethylation in the promoter of specific genes.
Next, we attempted to investigate the physiological significance of STAT1-TET2 complex mediated DNA demethylation. ALCL cells were known to turn into a drug-resistant state by long exposures of ALK tyrosine kinase inhibitors such as crizotinib. These drug-resistant ALCL cells exhibit STAT1 overactivation when the inhibitor is withdrawn, causing apoptosis1. Therefore, we hypothesized that STAT1 mediated demethylation induces drug resistance or cell apoptosis.
Knockdown of STAT1 did not influence the cell proliferation in ALCL cells, whereas it partially inhibited the sensitivity of crizotinib. Conversely, treatment of DNA demethylation agent 5-Aza or IFN-γ promoted the sensitivity of crizotinib. In addition, we generated drug-resistant ALCL cells by exposing crizotinib for an extended period of time. We confirmed these cells exhibited STAT1 activation when crizotinib was withdrawn, and drug-sensitivity recovered after a few days. We concluded that in the case of relatively weak STAT1 activation, ALCL cells increased drug sensitivity via demethylation.
Overall, our data suggests that activated STAT1 interacts with TET2 and STAT1-TET2 complex partially contribute to form DNA methylation pattern in ALCL cells and drug sensitivity (Figure 1.).
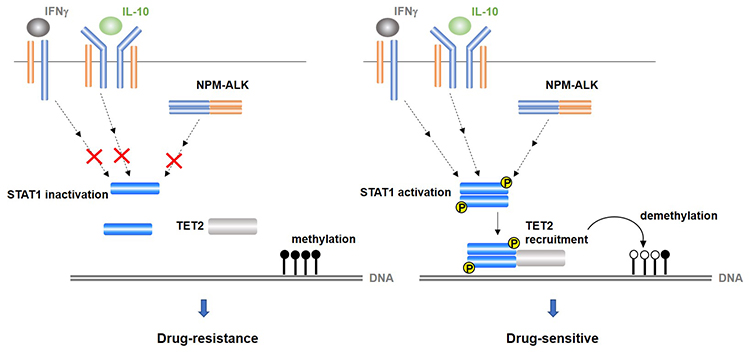
Figure1:
In the case of STAT1 inactivation, DNA methylation status remains stable and cells exhibit drug-resistance.
When STAT1 is activated by various stimulating factors, STAT1 interacts with TET2 and induces DNA demethylation at specific regions, causing drug-sensitivity.
Reference
- Rajan, et al. Oncogene. 2020 Mar 39 (10): 2103-2117.
Keio University Program for the Advancement of Next Generation Research Projects
The Keio University Program for the Advancement of Next Generation Research Projects subsidizes research costs with the aim of finding solutions to challenges and of promoting global academic research in order to allow Keio University faculty members to establish a presence as core researchers.