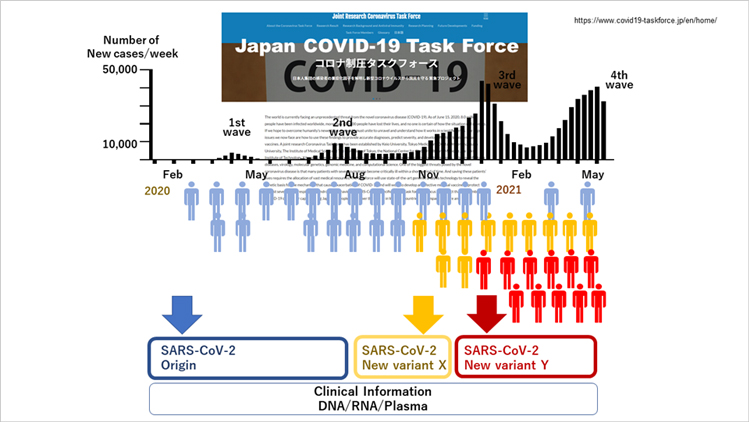
Since the COVID-19 pandemic, Japan COVID-19 Task Force has continued to collect clinical information and samples from patients during each virus variant epidemic and collaborated with more than 120 hospitals in Japan. © Japan COVID-19 Task Force
Keio University is leading a series of pioneering multidisciplinary projects combining clinical medicine with state-of-the-art genetic analysis of samples from COVID-19 cases for developing anti-viral drugs and improving the quality of life of patients.
Professor Koichi Fukunaga is a specialist in respiratory medicine at the Keio University School of Medicine in Tokyo. "I was recently asked to head Japan's Joint Research Coronavirus Task Force that was launched in May 2020 to investigate the spread and treatment of COVID-19 based on advanced genomic analysis," says Fukunaga. "We are focusing on trends in Japan. For example, we are looking into why the number of deaths in Japan has been significantly lower than in the United States and Europe. We have genetically analyzed thousands of blood samples taken at hundreds of hospitals throughout Japan in compliance with stringent ethics rules. We continue to share our findings from Japanese patients with researchers worldwide to contribute to global efforts to develop treatments to bring the COVID-19 pandemic to an end."
An example of the Joint Research Coronavirus Task Force's (Task Force) contribution to global research on COVID-19 includes sharing genomics data from Japanese patients with the Host Genetics Initiative as part of a project to identify the role of an individual's genetics in determining their "susceptibility and response to viral infection."1 The results of this global survey were published in the 8 July 2021 issue of Nature.2 "Our data set provided genetic information for the largest group in Asia that identified gene polymorphisms involved in the aggravation of coronavirus infection," explains Fukunaga. "Our study showed that Japanese individual's genetics information can affect their infection by COVID-19 and how it progresses. The results show the importance of our research activities in Japan."
Genome-Wide Association Studies
The membership and range of studies undertaken by the Task Force continue to evolve, with researchers from more than 120 hospitals across Japan, including 13 facilities affiliated with Keio University. As of October 2021, the Task Force had collected approximately 5,000 blood samples that are being analyzed according to the global standard procedures followed in Genome-Wide Association Studies (GWAS).
The process of analysis is as follows. Samples are grouped into DNA/RNA, plasma, and blood cells, and sent to specialists in the Task Force for analysis. For example, multiomics and GWAS analysis are carried out at the Tokyo Medical and Dental University and Kyoto University; immunology analysis at Keio University; and data management at Tokyo University's Institute of Medical Science. Next, the data from this analysis is sent to researchers at Osaka University to identify risk factors, and specialists at Keio University and Kitasato University for 3D organoid assay analysis. "The platform that we have established for integrating the research from the hundreds of members of the Task Force enables the control of the flow and management of not only genetics data, but also of clinical information, which is a critical feature of this project," says Fukunaga.
Clinical research activities on COVID-19 at Keio University: K-CORC
"In addition to our activities as part of the Task Force, researchers at Keio University are also conducting independent research on COVID-19 to clarify the clinical features and risk factors for people who were infected with COVID-19 by community transmission," explains Fukunaga. "The Keio COVID-19 Research Consortium (K-CORC) consists of Keio University Hospital and 13 community hospitals located in and around Tokyo and has been in operation since the first COVID-19 wave in Spring 2020. We have already collected data on 345 patients admitted for COVID-19 infection from February to June 2020." Notably, the results for the approximately 32% of patients requiring administration of oxygen showed that the main risk factors were 'consciousness disorder,' older age, high blood pressure, diabetes, and a history of heavy smoking.3 Furthermore, the main factors leading to death were chronic kidney disease and hyperuricemia/gout.
Post-genomics, 'Factor-X', and blood types
"Statistics from the spring of 2020, when COVID-19 began to spread globally, showed that the fatality rate from COVID-19 in Japan was much lower than in the US and Europe," says Fukunaga. "This led scientists in Japan to consider the possibility of an 'X-Factor' related to the genetic make-up of Japanese people that kept the death rate low here. One of the roles of the Task Force was to try to clarify if there is indeed a link between COVID-19 deaths and genetics in Japan."
To resolve this issue, researchers in the Task Force conducted GWAS analysis of approximately 2,393 COVID-19 cases and 3,289 controls, and, importantly, identified that the DOCK2 gene may be a factor in severe cases of COVID-19 in Japan for patients who were less than 65 years old.4 "This was the largest ever GWAS-based study regarding COVID-19 on Japanese people, and we have shared our results with international groups," says Fukunaga. "Furthermore, our more recent GWAS clinical studies on patients aged less than 65 showed blood group to be an important factor for severe COVID-19 cases. We found AB-type patients were the highest risk at 40% and O-type the lowest at 20%. Obesity and hyperuricemia/gout are also high-risk factors for Japanese groups. These findings were only possible because we combined results from genetic analysis and hands-on clinical data of patients throughout the COVID-19 pandemic."
Furthermore, genetic studies of Japanese patients revealed the existence of a single-nucleotide polymorphism (SNP) on chromosome 5 in 5q35 genes with G/A mutation 5q35.1. Notably, G mutated to A in 84.1% of patients who were less than 65 years of age; this is a 1.6-fold increase in this mutation in severe COVID-19 cases. These results show that nearby variants (in this case G mutating to A) can affect mRNA transcription (protein expression). A candidate for this gene is the DOCK2.
The DOCK2 gene was identified as playing a major role in severe cases of COVID-19.4 Also, single-cell RNA sequencing of blood samples from COVID-19 patients showed a decrease in DOCK2 expression in the monocyte region. These results confirmed that inhibition of DOCK2 exacerbated COVID-19 pneumonia in an experimental Syrian hamster model.
"We continue to share our results with the global community of COVID-19 researchers," says Fukunaga. "The recent paper in Nature was based on data correlated by members of the COVID-19 Host Genetics Initiative, and the sharing of our data was thanks to the Host Genetics Initiative."
High throughput screening with organoid based assay
In a project led by Toshiro Sato at the Keio University School of Medicine, Keio researchers are laying the foundations for high throughput screening with 3D organoid-based assays. Specifically, the Keio team has established procedures to cultivate 3D organoids for human airway epithelium and alveoli.5 "Our ability to cultivate 3D organoids that replicate human tissue is important because it will enable us to undertake trials of drugs to treat COVID-19," explains Fukunaga. "The 3D organoid-based experiments are far more powerful in giving us deeper insights into the treatment of viral diseases than hamster- or mouse-based trials. The simple reason is that the organoids are 3D tissue structures cultivated using actual human cells that give direct feedback about effects of drugs on such diseases."
Future plans: Post-COVID conditions and quality of life
The spread of COVID-19 has led to mutations that continue to pose threats to society worldwide. In Japan, the characteristics of the first and fourth waves were different as the virus mutated and spread throughout the community. "To date, we have looked at COVID-19 from the patient's perspective," says Fukunaga. "In the future, we want to use all the data that we have accumulated to-date to look for trends that may give signs about the possibility of mutation and other changes about the behavior of the virus." To achieve these goals the Task Force is setting up a comprehensive platform to collate, share, and distribute clinical and genetic information on COVID-19 with the goal to develop next generation drugs. "This is a major undertaking necessitating extensive manpower, funding, and management skills," says Fukunaga.
The majority of people who are infected by COVID-19 recover within a few weeks. However, others experience so-called 'post-COVID-19 conditions' even after several weeks since first being infected. Fukunaga and colleagues at Keio University are studying this aspect of the COVID-19 pandemic as well. "We are working on a project funded by Japan's Ministry of Health, Labour and Welfare on clarifying the frequency, types of symptoms, and longevity of post-COVID conditions in patients," explains Fukunaga. The project involves 28 medical facilities in Japan, with Keio acting as the managerial hub for disseminating funding and collecting results. The project ends in March 2022 and the team plans to collect 1000 samples from patients showing mild, medium, and severe symptoms. "We are looking at patients from the date of admission to 3 months, 6 months, and in some cases 12 months afterward" Fukunaga says. The project includes industrial partners who are responsible for developing technology such as the 'e-PRO'―an electronic reporting app―for collecting patient-reported outcomes and the EDC, or 'Electronic Data Capture,' system.
"We have obtained extensive data on symptoms such as 'brain fog' and hair loss. These findings are valuable for putting COVID-19 patients at ease about their symptoms, where we can tell them about the extent and longevity of similar symptoms experienced by other people based on real first-hand data. I am convinced the combination of clinical and genetic information is important for improving the quality of life of people suffering from COVID-19 and for mitigating such emergencies in the future. This truly encapsulates the ultimate goals of the Task Force."
Published online 28 January 2022
About the researcher
Koichi Fukunaga M.D, Ph.D ― Professor
Department of Internal Medicine, School of MedicineKoichi Fukunaga was born in Tokyo, Japan, in 1969. He is a graduate of Keio Senior High School and the Keio University School of Medicine. In 2019, he started working at the Pulmonary Division of the Keio University Department of Medicine as a professor. He was also appointed as Vice Director of Keio University Hospital in 2021. His research interests are in developing new strategies for inflammatory lung diseases.
Links
Reference
- Host Genetics Initiative, accessed 21 December 2021.
- Initiative, C.-19 H. G. Mapping the human genetic architecture of COVID-19. Nature 600, 472-477 (2021). | article
- Ishii, M. et al. Clinical characteristics of 345 patients with coronavirus disease 2019 in Japan: A multicenter retrospective study. J.Infect. 81, e3-e5 (2020). | article
- Namkoong, H. et al. Japan COVID-19 Task Force: a nation-wide consortium to elucidate host genetics of COVID-19 pandemic in Japan. medRxiv 2021.05.17.21256513 (2021). | article
- T. Ebisudani, S. Sugimoto, K. Haga, A. Mitsuishi, R. Takai-Todaka, M. Fujii, K. Toshimitsu, J. Hamamoto, K. Sugihara, T. Hishida, H. Asamura, K. Fukunaga, H. Yasuda, K. Katayama, T. Sato, Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening. Cell Rep. 35, 109218 (2021). | article