Cutting edge mass spectrometry technology offers unprecedented insights into possible links between the functional metabolites produced by gut microorganisms and human health
Lipids are hydrocarbon molecules―such as fats and oils―that form the building blocks of human cell membranes and store energy that is released by a variety of metabolic pathways for use by the body. "The extremely diverse nature of lipids means that it is not only challenging to identify and determine species but also to quantify their roles as signaling molecules in physiology and disease," says Makoto Arita, a professor at the Keio University Faculty of Pharmacy. "My research is focused on gaining insights into the structure and functions of lipids that govern inflammation and tissue homeostasis.1 My approach is based on the analysis of lipids using liquid chromatography tandem mass spectrometry (LC-MS/MS)―an extremely powerful tool for lipidomics research that we have recently been using to elucidate a wide range of functional metabolites produced by gut microorganisms."
Advanced non-targeted lipidomics for gut microorganisms
Professor Arita recently reported on the development of new lipidomics technology to analyze complex lipid metabolites produced by gut microorganisms in collaboration with colleagues at RIKEN, where he is the team leader of the Laboratory
for Metabolomics at the RIKEN Center for Integrative Medical Sciences.2,3 "The new lipidomics technology combines non-targeted mass spectrometry to enable unbiased comprehensive analysis with feature-based molecular spectrum networking, which
supports structural estimation of unknown molecules," explains Arita.
The research team used mice treated with different combinations of antibiotics to successfully identify 985 lipid structures in feces, of which around 24.8% appeared to be dependent on gut microorganisms as determined by a more than 10-fold
decrease in antibiotic-treated mice. They also identified unique lipid structures that are directly related to gut microorganisms, including acyl alpha-hydroxyl fatty acid (AAHFA). Notably, this approach accurately predicted the bacterial species
responsible for producing AAHFA and other unique lipid structures.
"We expect this research to lead to unprecedented insights into the complex lipid metabolism networks resulting from the interaction between gut microorganisms and their hosts," says Arita. "Ultimately, this research could potentially
lead to the discovery of new functional metabolites involved in host homeostasis and the onset of diseases."
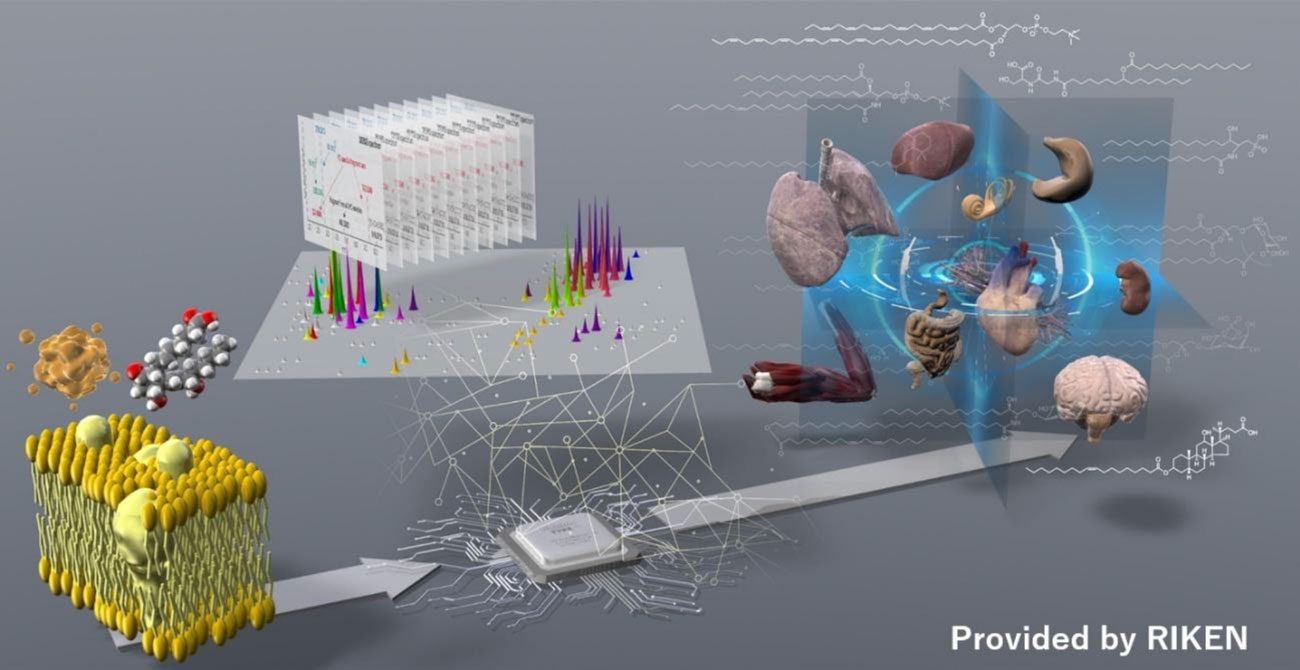
Non-targeted lipidomics to address the biological questions © RIKEN
Published online 14 July 2021
About the researcher

Makoto Arita ― Professor
Division of Physiological Chemistry and Metabolism,Faculty of PharmacyMakoto Arita received his Ph.D. from the University of Tokyo in 1997. Since 2014, he has headed the Laboratory for Metabolomics at the RIKEN Center for Integrative Medical Sciences. In 2016, he became professor at the Keio University Faculty of Pharmacy. His research interests are in the molecular mechanisms through which specific lipids elicit their biological functions involved in health and diseases.
Links
Reference
- H. Tsugawa et al., A lipidome atlas in MS-DIAL 4, Nat Biotechnol 38, 1159-1163, (2020). | article
- S. Yasuda et al., Elucidation of Gut Microbiota-Associated Lipids Using LC-MS/MS and 16S rRNA Sequence Analyses, iScience, 23, 101841, (2020). | article
- N. Okahashi et al., Global profiling of gut microbiota-associated lipid metabolites in antibiotic-treated mice by LC-MS/MS-based analyses, STAR Protocols, 2, 100492, (2021). | article