Dynamic interactions between proteins and lipids at cell membranes result in complex changes in diffusion rates
Published online 29 June 2017
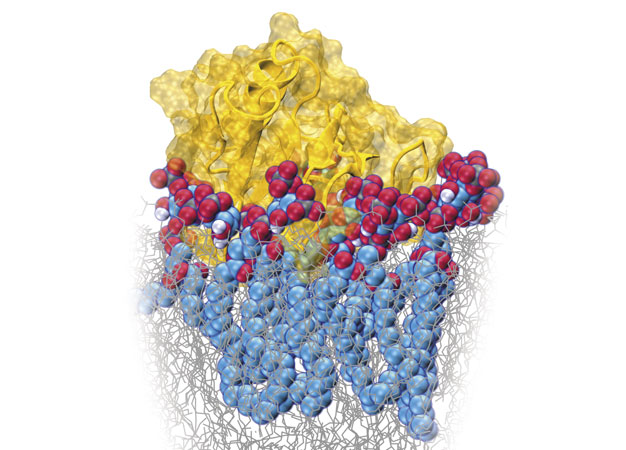
Molecular dynamics simulation snapshot showing the binding of a Pleckstrin homology domain (yellow) with lipids (red/blue) in the cell membrane (grey).
Reproduced from Ref. 1 and licensed under CC BY 4.0 © 2017 E. Yamamoto et al.
Molecular dynamics simulations have revealed that the diffusion of key biomolecules at cell membranes changes dramatically over short time scales due to complex molecular interactions, including clustering1. This suggests that the apparently steady diffusion of these molecules over longer time scales hides a much more complex molecular 'walk', which may be a critical component of biological processes.
The cell membrane is a remarkable biological structure that is selectively permeable to certain ions and biomolecules while being impermeable to most water-soluble molecules. Peripheral membrane proteins (PMPs) temporarily bind to the cell membrane to provide a range of biological functions, including the regulation of cell signaling and other important cellular events. As many of these functions are determined by the speed at which PMPs approach, attach and detach from the membrane, an understanding of the detailed molecular dynamics of these processes could provide important information on biological mechanisms. However, conventional methods for modeling and analyzing the diffusion of PMPs provide only general characteristics and are unable to resolve the detailed motion contributing to overall diffusion over long periods.
Kenji Yasuoka and colleagues from Keio University, in collaboration with researchers from the University of Oxford and University of Leeds in the UK, have now used high-resolution molecular dynamics simulations to show that there is much more to the story.
"We found that the diffusivity of the protein fluctuates anomalously due to dynamic interactions between the protein and specific lipids," explains Yasuoka. "This 'short-time' diffusivity is intrinsically different from long-time measurements, and shows that the overall mobility of signaling proteins on the surface of cell membranes is sensitive to lipid interactions."
The researchers used a new method to estimate the short-time diffusivity from a single molecular trajectory without relying on knowledge of changes in diffusivity, looking specifically at the dynamic interactions of the Pleckstrin homology (PH) domain ― the PMP lipid recognition module that controls membrane binding. Their molecular dynamics simulations tracked the motion of a hundred Pleckstrin homology domains when placed within a few nanometers of a cell membrane surface over a period of just 10 microseconds.
The results show that cell membranes are complex spatially and temporally inhomogeneous environments where crowding of lipids and proteins is common.
"Determining the interactions of PMPs with the membrane at the molecular level is crucial for understanding the function of these biological important proteins," says Yasuoka. "We are also interested in investigating the diffusion process in more complex systems, such as the interaction between transmembrane proteins and PMPs."
Reference
- Yamamoto, E., Akimoto, T., Kalli, A. C., Yasuoka, K. & Sansom, M. S. P. Dynamic interactions between a membrane binding protein and lipids induce fluctuating diffusivity. Science Advances 3, e1601871 (2017). | article