The influence of intestinal bacteria on gene expression in immune cells could be the key to treating inflammatory bowel disease
Published online 10 September 2015
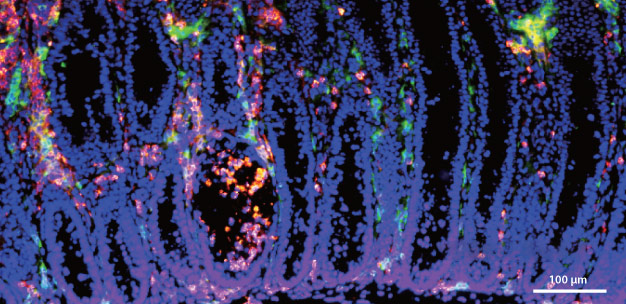
Disrupting the epigenetic regulation of regulatory T cells in the intestinal lining of mice causes inflammation, shown by the green fluorescent markers.
Reprinted by permission from Macmillan Publishers Ltd: Nature Immunology Ref. 3, copyright 2014
The past 100 million years of evolution have developed a complex relationship between bacteria and the human gut. These bacteria are essential because they break down nutrients that we cannot process, but, conversely, our immune systems must be constantly alert to the possibility that they will trigger an inflammatory response. Rapidly unfolding research that includes cutting-edge work at Keio University is revealing the importance of epigenetic mechanisms to this interplay, in which intestinal bacteria influence gene expression in immune cells. The findings present novel opportunities for the treatment of inflammatory bowel disease, which affects more than 3.5 million people worldwide.
Epigenetics and immunity
Epigenetic mechanisms involve chemical modifications of DNA molecules and histones, the proteins around which they are wrapped. These modifications alter gene expression and, consequently, cell function. The significance of epigenetics was described by the British biologist and geneticist, Conrad Waddington, as early as 1942, but the mechanisms and their full implications are still being unraveled.
"Nowadays, human epigenetic studies are common," says Koji Hase from the Keio University Graduate School of Pharmaceutical Science. The US National Institutes of Health (NIH), for example, launched the Roadmap Epigenomics Mapping Consortium in 2008, which has since revealed the epigenomic landscape in various human tissues and cells, and could lead to state-of-the-art therapies for cancers and autoimmune diseases. "Epigenetics in the immune system, however, is an emerging research field that has rapidly developed since the turn of the century," says Hase.
Since then the crucial role of epigenetics in the immune system has become clear1. For example, histone modifications alter the function of macrophages ― cells that ingest dead and dying cells, as well as pathogens ― and studies in mice have shown that abnormal modifications are associated with diseases such as helminth infections and Parkinson's. In 2014, an international study established that disrupted modification of DNA in cells that line the intestine can cause colorectal cancer.
Immunity to inflammation
Research at Keio University, undertaken by several scientists, has focused on the epigenetic effects of intestinal bacteria on immune cells. "Keio University is now the center of microbiome and epigenetic studies in Japan, and researchers here are leading the worldwide initiative in microbiome studies," says Hase. Notable researchers include Kenya Honda, who heads research on the regulatory effects of the microbiome on the immune system, Takanori Kanai, who was the first to initiate fecal microbiota transplantation in Japan, and Akihiko Yoshimura, who leads studies on the epigenetic regulation of immune cells.
Hase's own work has shown that a metabolite called butyrate that is produced by intestinal bacteria is involved in the epigenetic regulation of regulatory T cells2,3, which are central to the suppression of inflammatory responses. Butyrate was found to induce the differentiation of regulatory T cells in mice. Disruption of this epigenetic regulation caused severe inflammation of the intestinal lining known as colitis (see image)3, but the disease was prevented by treatment with butyrate2.
"Our findings not only link butyrate to microbe-mediated induction of regulatory T cells in the colon, but also provide evidence for the therapeutic application of butyrate in inflammatory bowel disease," says Hase.
Although the Keio University research has established the importance of intestinal bacteria in the epigenetic regulation of immune function, the precise molecular mechanisms behind this regulation are not known. Determining these mechanisms is Hase's next step.
"I think that gut-derived metabolites might behave as 'external hormones' to regulate host immunity and metabolism," he says. "I would like to take advantage of metabolomics to identify the molecular components of such host-microbe signaling."References
- Obata, Y., Furusawa, Y. & Hase, K. Epigenetic modifications of the immune system in health and disease. Immunology and Cell Biology 93, 226-232 (2015). | article
- Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446-450 (2013). | article
- Obata, Y. et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nature Immunology 15, 571-579 (2014). | article